Abstract
Believe it or not, scientists were recently able to recover tissue from a 68-million-year-old Tyrannosaurus rex fossil! Not only were they able to purify non-mineralized tissue, but they also succeeded in obtaining partial sequence information for protein molecules in the T. rex tissue. In this genomics science fair project, you will use the T. rex's protein sequence to search sequence databases for the its closest living relatives.Summary
David B. Whyte, PhD, Science Buddies
Edited: Svenja Lohner, PhD, Science Buddies

Objective
The objective of this genomics science fair project is to determine the closest living relative to the mighty Tyrannosaurus rex, using simple bioinformatics tools.
Introduction
Have you ever noticed that birds have scales on their feet? The reason they have scales is that, technically speaking, they are reptiles, and reptiles have scales. What about the feathers? Feathers are produced by tissues similar to those that produce scales. Also, birds lay eggs like other reptiles. Not only are birds considered reptiles, but scientists now generally agree that birds are, in fact, dinosaurs. Specifically, birds are members of the clade Maniraptora (a clade is a group of animals related by descent from a common ancestor). Maniraptorans all have shared skeletal features, including bone structures in the wrist and forelimb that were first used for grasping, but that were modified into wings during the evolution of birds.
The Maniraptora is the group of theropod dinosaurs. The major Maniraptoran groups include:
- Aves: The birds, living dinosaurs.
- Dromaeosaurs: The "raptors," including velociraptor, made famous in the movie Jurassic Park.
- Troodontids: Non-avian dinosaurs thought by some to be particularly intelligent.
- Therizinosaurs: Plant-eating theropods.
- Oviraptors: The fossil record contains evidence that these dinosaurs were devoted parents.
It is important to note that birds are not descended from velociraptors or any of the other maniraptorans. They are all derived from a common ancestor. Birds split from the other members of the group about 150 or so million years ago, in the Jurassic period. The non-avian dinosaurs became extinct over 65 million years ago, but the birds have flourished.
The evidence that birds are dinosaurs is based on detailed studies of fossils, as well as the biology of modern birds. Recently, a new avenue of analysis became available with the extraction of tissue from dinosaur bones. Dr. John Asura, and other scientists, published an account of their analysis of collagen proteins purified from bones of a Tyrannosaurus rex (T. rex) in the journal Science, which you can find in the Bibliography section below. They were able to obtain partial sequence information from the T. rex collagen proteins. Although the protein sequence they obtained is not complete (see the Procedure section for the actual sequence), it has enough information to allow searching of sequence databases.

Figure 1. T. rex head reconstruction at the Oxford University Museum of Natural History. (Wikipedia, 2006.)
BLAST is a program used to search databases of sequence information. For this science fair project, you will search SwissProt, a database of protein sequences. Each record has the protein sequence, as well as the authors who submitted the sequence, the article associated with the sequence, and other information.
In the Procedure, you will use BLAST to search the SwissProt protein database for sequences related to the T. rex sequence. If two organisms are descended from a recent common ancestor, their protein sequences will be similar. For example, the collagen genes in two species that split 1 million years ago will have fewer differences than two species that split 10 million years ago. This is because DNA accumulates mutations over time. If the rate at which mutations accumulates is constant, the number of mutations is proportional to the time since the species split. The mutations that are accumulated over time are useful in phylogenetic analysis. You might ask, what is phylogenetic analysis and how are mutations used? Phylogenetic analysis simply means the study of evolutionary relationships among organisms. Mutations play a role in phylogenetic analysis because they are ideal "tags" for lines of descent. The presence of a particular mutation within a group of animals is evidence of a common ancestor. Most mutations are not passed down to subsequent generations, but some do become common within the population. Based on the differences in DNA or protein sequences, one can create a phylogenetic tree, which is a tree-like diagram showing the evolutionary relationships among different biological species based upon similarities and differences in their genetic characteristics. In other words, you can use protein or DNA sequence comparisons to establish how animals are related to each other. You can watch the two videos below to learn more about phylogenetics and reading phylogenetic trees.
Using BLAST, and publicly available databases, you can perform your own genomics science fair project, studying the evolutionary relationships of various animals. Now that the database contains sequence information for T. rex, you have the tools needed to investigate which of the organisms represented in the SwissProt database is most related to this extinct dinosaur.
Terms and Concepts
- Clade
- Maniraptora
- Evolution
- Therapod
- Ave
- Dromaeosaur
- Troodontid
- Therizinosaur
- Oviraptors
- Collagen proteins
- BLAST
- Mutation
- Phylogenetic analysis
- Phylogenetic tree
- Query
- Fasta format
- Phylogenetic tree
- Evolution
Questions
- What does the acronym BLAST stand for?
- Based on your research, draw a family tree that includes birds, dinosaurs, reptiles, and mammals.
- What dinosaurs have been found to have feathers?
Bibliography
These websites offer more information about dinosaurs, specifically those discussed in this science fair project:
-
Asara, J.M., et al. (2008, April 25).
Molecular Phylogenetics of Mastodon and Tyrannosaurus rex.
Science, Vol. 320., No. 5875, p. 499. Retrieved August 25, 2008, from National Center for Biotechnology Information.
- PDF copies of the Molecular Phylogenetics of Mastodon and Tyrannosaurus rex article and the supplementary data.
- Vergano, D. (2007, April 12). Yesterday's T. Rex is today's chicken. USA Today. Retrieved August 25, 2008.
- DinoBuzz, University of California Museum of Paleontology. (n.d.). Are birds really dinosaurs?. Retrieved August 25, 2008.
- DinoBuzz, University of California Museum of Paleontology. (n.d.). Maniraptora. Retrieved August 25, 2008.
These websites are useful resources for understanding how DNA can be used to build evolutionary trees and the bioinformatics tools used to do this:
- National Center for Biotechnology Information. (2008). NIH Home. Retrieved August 25, 2008.
- O'Halloran, D. (2014). A Practical Guide to Phylogenetics for Nonexperts. Journal of Visualized Experiments, Vol. 84, 50975. Retrieved September 3, 2021.
Materials and Equipment
- Computer with access to the Internet
- Lab notebook
Experimental Procedure
The procedure for this genomics science fair project has two sections: 1) Use BLAST to search SwissProt (an online database of protein sequences) for the best match to Tyrannosaurus rex sequence data (a query sequence is used to search the database), and 2) Build a tree graphically showing the relationship of T. rex to its living relatives.
Before you start with this project, it might be helpful to familiarize yourself with the bioinformatic tools that you are going to use. You can watch the video below to learn more about the BLAST tool on the NCBI website.
Use BLAST to Search SwissProt
-
The partial sequence for the Tyrannosaurus rex collagen protein is pasted below. It is from the Science article by Asara, listed in the Bibliography at the end of the Background section. Regions where the protein sequence is not known have a hyphen (–) to represent a gap of indeterminate length. The capital letters each represent an amino acid in the protein sequence. Note that most of the protein was not successfully sequenced, but considering that the tissue was 68 million years old, it is remarkable any sequence was obtained. The protein sequence is in FASTA format, which means that the sequences are preceded by a header line that starts with a ">" and ends with a return, or a new paragraph. The FASTA format is the standard formatting used by bioinformatics software.
>Tyrannosaurus rex, collagen type I, alpha 1 -GATGAPGIAGAPGFPGARGAPGPQGPSGAPGPK-GVQGPPGPQGPR-GSAGPPGATGFPGAAGR-GVVGLPGQR-GLPGESGAVGPAGPPGSR-
- Copy the sequence of the T. rex collagen protein above, including the header line (> Tyrannosaurus rex) and all of the hyphens.
- Open a BLAST page at the National Center for Biotechnology Information (NCBI).
- Go to the NCBI main page.
- Click on the BLAST link in the "Popular Resources" list on the right to get to the BLAST page
- There are several versions of BLAST. Since you want to use a protein sequence to search a protein database, click on "Protein BLAST" under the "Web BLAST" heading.
- Fill out the protein BLAST query form, which should look similar to Figure 2, below, when you are done.
- Paste the T. rex sequence into the "Enter Query Sequence" box.
- For job title, use "Tyrannosaurus rex, collagen type 1, alpha... ."
- If you kept the header line (>Tyrannosaurus rex) at the top of the sequence, it will be added here automatically.
- Under "Database," choose the "UniProtKB/Swiss-Prot(swissprot)" protein database.
- Leave the box for "Organism" empty.
- If you would like to compare the genomes of animals other than the default ones, you could add them here by entering their names.
- Under "Algorithm," select blastp (protein-protein BLAST).
- Next to the BLAST button, check the box "show results in a new window."
- Click on "Algorithm Parameters" underneath the BLAST button. In the "General Parameters" section, select 10 for the Max target sequences. This will limit your search to the closest 10 protein sequences and simplifies your phylogenetic tree in the following step.
- You can expand your search to 50 or more target sequences later and also explore other BLAST options in the "Algorithm parameters" section.
- Then click on BLAST to start the search.
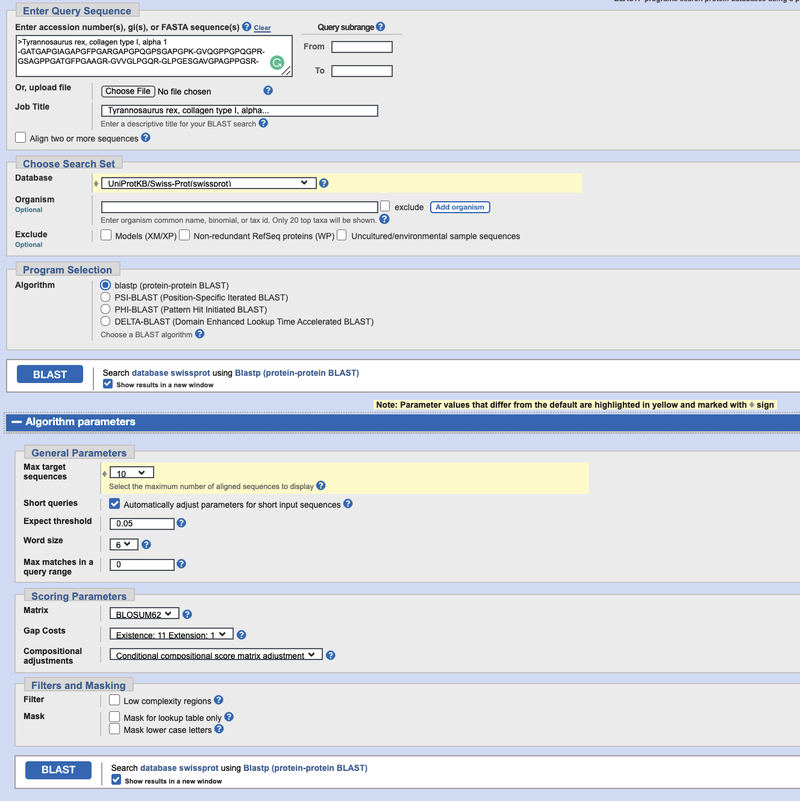 Image Credit: (BLAST) NCBI main page / Public Domain
Image Credit: (BLAST) NCBI main page / Public DomainScreenshot of the search page on the ncbi.nlm.nih.gov website. At the top of the BLAST query search page there is a text box where users can fill in search terms. Other options are available under the search box that allow for different databases to be searched and to limit searches through keywords or IDs.
Figure 2. Protein BLAST (blastp) query input page. Your query page should look similar to this one after you have filled it in by following step 4, above.
- Be patient. It will take a few minutes for the BLAST results to appear. See Figure 3, below, for a snapshot of how the results page should look like.
- On the top left of the BLAST results page you will find the summary section (blue in Figure 3), which provides information on different aspects of your search. On the top right there is a box that allows you to filter your results based on certain criteria (red in Figure 3). Below the top section, the BLAST results are shown (yellow in Figure 3). There are four different tabs called "Description," Graphic Summary," "Alignments," and "Taxonomy." Each tab presents the search results in a different way.
- The "Description" tab contains a summary table of hits found by BLAST and is the default tab shown.
- The "Graphic Summary" tab shows a color key of the alignments. The color key shows the degree of similarity for the sequences.
- The "Alignment" section contains the detailed pairwise alignments between query and database sequences.
- The "Taxonomy" section provides details of the taxonomic distribution of matches BLAST found.
- Review each of the four tabs. Based on the information provided, can you tell what living organism is most related to T. rex, based on similarity of collagen genes?
- Scroll down the the result list in the "Description" tab. Note the "Max Score" column. Proteins with the highest scores are most related to the T. rex query sequence. The E value is an estimate of the chance that the sequences are not related. The smaller the E value, the more certain the sequences are related. You will also find the scientific names of each species that matched to the T. rex collagen sequence in the "Scientific Name" column.
- In the"Alignments" tab you will find the alignments of the T. rex amino acid sequence with the sequences in the database. Note the "Identities" value, which is the percent of amino acids that are the same in the query and the database sequence. "Positives" measures the percent of amino acids that remain the same or that were changed into similar amino acids. If the % identity between two species is 97%, then these two species differ by 3% in the protein sequence. Remember, the larger the % difference, the more distant they are in the family tree.
- Make a data table based on the BLAST output. List the organism's scientific name, common name, the score, % identity, and the E value. View each of the four result tabs to find the information you need.
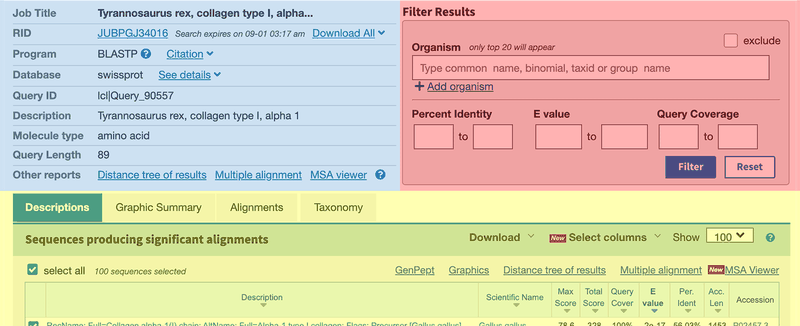 Image Credit: (BLAST) NCBI main page / Public Domain
Image Credit: (BLAST) NCBI main page / Public DomainScreenshot of the results page in the BLAST tool on the ncbi.nlm.nih.gov website shows a list of protein sequences that match a search term. Results provide additional information such as the percentage match a result has to the specific query string that was searched for.
Figure 3. Snapshot of the BLAST outout page.
Make a Phylogenetic Tree
In this section, you will use the BLAST output to make a tree that graphically depicts the degree of similarity of the proteins. There are more sophisticated ways to generate a phylogenetic tree, which you can explore in the variations in the Make It Your Own section.
- To generate a tree of the BLAST results, click on "Distance tree of results" next to "Other reports" above the results table in the summary section of your BLAST report. This tree includes all of the hits from the BLAST search.
- The tree will be displayed in "rectangular" format.
- Your query sequence will be higlighted in yellow.
- Keep all the default setting for the tree parameters but change the "Sequence Label" to "Taxonomic Name" in the drop-down list.
- Add the common name for each species to the tree.
- This might be easier if you redraw the tree by hand or use a computer graphics program.
- Add BLAST data, such as the % identity to the tree.
- What does the phylogenetic tree tell you about the closest living relatives of T. rex? If you need help reading the phylogenetic tree, you can view the Phylogenetics and Reading Phylogenetic Trees or the Phylogenetic analysis of pathogens video.
Ask an Expert
Variations
-
Build a phylogenetic tree based on the sequences used in the original paper by Asara, et al.
- First, download the sequences for the collagen genes that were used in the Asara paper, Molecular Phylogenetics of Mastodon and Tyrannosaurus rex.
- Open the ClustalW2 tool at the EBI Bioinformatics site (also referenced in the Bibliography).
- Paste the sequences into the entry box and click "Run."
- After the alignment is complete, click on "Jalview" for a set of tools for making various trees.
- Add pictures of the animals to the appropriate branches of the phylogenetic tree.
Careers
If you like this project, you might enjoy exploring these related careers:
Related Links
- Science Fair Project Guide
- Other Ideas Like This
- Genetics & Genomics Project Ideas
- Big Data Project Ideas
- My Favorites